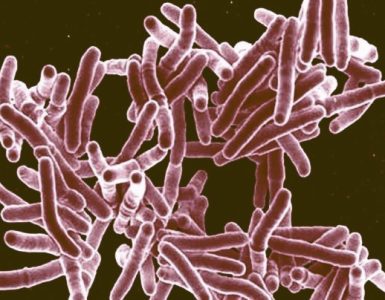
Following previous efforts, BGI and their collaborators at the University Medical Centre Hamburg-Eppendorf, as well as a growing number of researchers around the world “crowdsourcing” this data, are exploring in-depth the European disease outbreak helping trace the origin and spread of the lethal E. coli strain. Different sources have reported that two strains, 01-09591 from Germany isolated in 2001 and 55989 from Central Africa in 2002, are highly similar to the 2011 outbreak strain. Based on a the most recently curated assembly publically released by the BGI yesterday (ftp://ftp.genomics.org.cn/pub/Ecoli_TY-2482), these strains have an identical Multi Locus Sequence Typing (ST678) based on analysis of 7 important “housekeeping” genes*.
The relevance of this data is we are now tracing the history of the bacteria, as this latest analysis indicates that the two German strains (01-09591 originally isolated in 2001 and TY2482 from the 2011 outbreak) have identical profiles for all 12 virulence/fitness genes and 7 MLST housekeeping genes. However, at some point over this 10-year period the new 2011 outbreak strain seems to have developed the ability to resist many additional types of antibiotics. The latest data is now pointing to this candidate, as it now seems the African strain (strain 55989) is genetically more “distant” as the Shiga-toxin-producing gene and tellurite-resistance-genes were shown to be absent. (see ftp://ftp.genomics.org.cn/pub/Ecoli_TY-2482/2011vs2001_v2.xls for our detailed comparison). The utility of so quickly sharing our initial data is further supported as the link to this original strain has already been independently verified by other groups: http://scienceblogs.com/mikethemadbiologist/2011/06/i_dont_think_the_german_e_coli.php). See also ColiScope where the sequence of strain 55989 was first displayed (https://www.genoscope.cns.fr/agc/microscope/mage/viewer.php, with option chromosome EC55989_EC55v2)
This latest evidence that the previously 2001 German strain is the most likely ancestor of the 2011 outbreak strain. This may imply that fast evolution resulted in the gain of more genes during the last 10 years. Further comparisons between the genomes of these bacteria will greatly help clarify why the latest outbreak has been so exceptionally pathogenic on this occasion, and also provide clues on tracing the origin, spread and source of the disease, which would significantly aid the frontline health care workers fighting to control this now global outbreak.
Unfortunately, the 2001 German strain currently has no publically available genome sequence, although it was preliminary analyzed during the original outbreak and stocks and samples are hopefully still stored. In the great strides already made by the community in just a few days from the sharing of our original genomic data, we at the BGI is appealing for any labs who have isolates of this key strain to share samples and respective data for sequencing and analysis. The idea seems to be welcomed by scientific community. “The origin (patient index) of the E. coli EHEC O104:H4 is not yet known. One may however remember that a strain with a similar surface antigen was isolated in Germany in 2001.” Antoine Danchin (Microbiologist and founder of the HKU-Pasteur Research Centre) further stresses this in a recent posting: (http://www.normalesup.org/~adanchin/populus/journalist.html ), “Knowing the kinship between the present strain and the older one will be of the utmost importance.”
This comparative work is now of the upmost urgency, so please contact us caosujie@genomics.org.cn if you would like to collaborate in this effort by contributing samples or data from strain HUSEC041/01-09591.
The international community “crowdsourcing” and analyzing our most up-to-date genomic data now has a repository to keep this preliminary analysis together, and we would recommend scientists use our latest improved assembly (http://www.ncbi.nlm.nih.gov/bioproject/67657) and follow the repository and our Twitter feed (@BGI_Events) for the most up-to-date announcements and data:https://github.com/ehec-outbreak-crowdsourced/BGI-data-analysis
*Technical note: while there have been some reports that the sequence of MLST genes of 2001 Germany isolate and the 2011 outbreak strain may have minor differences
(http://scienceblogs.com/mikethemadbiologist/2011/06/i_dont_think_the_german_e_coli.php), BGI bioinformatician believe these are likely discrepancies mainly due to sequencing and assembly errors. Using the most recently curated assembly from BGI, we have found that the three strains do have identical MLST. There is also likely confusion from previous reports doing MLST comparisons with a previous US isolate with serotype O107. We have now found this US isolate to be significantly different from the Germany 2001 isolate, the former having all 7 MLST genes different from the outbreak strain although having the same serotype.
Source: BGI