Credit: Veronica Falconieri, Sriram Subramaniam, National Cancer Institute, National Institutes of Health
In 1977, Carl Woese et al introduced the three-domain system of biological classification that divides life forms into Archaea, Bacteria and Eukaryotes. This was the first time that the differences between Archaea and Bacteria were recognized. The first observed archaea were extremophiles, able to survive and thrive in extremely harsh conditions, but ongoing research has found them in a wide range of habitats; they are particularly abundant in the oceans. Archaea form part of the microbiota of all organisms, including humans, and their unique survival capabilities mean they can be used to enhance our understanding of early life on Earth. We are just beginning to investigate viruses that can infect Archaea, and in work recently published in Nature Communications, an international team of researchers determined the near-atomic structures of HCIV-1 and HHIV-2 by cryo-electron microscopy (cryo-EM).
Their results show how nature has solved the complexity of arranging vertical single β-barrels and provide insights into the evolutionary consequences of the fusion event of the two consecutive major capsid protein (MCP) genes, which has ultimately led to the membrane-less vertical double β-barrel MCP assembly seen in adenovirus.
Complex self-assembly
Professor Nicola Abrescia at the CIC bioGUNE (Spain) and his colleagues Prof Bamford and Dr. Oksanen at the University of Helsinki enjoy poking around in salt pans and other unexplored environmental niches, combining fishing trips to collect new viruses with work or leisure trips around the world. This work investigates two related viruses, membrane-containing Haloarcula californiae icosahedral virus 1 (HCIV-1) and membrane-containing Haloarcula hispanica icosahedral virus 2 (HHIV-2), samples of which were collected in Thailand and Italy, respectively. Initial research to identify the host and virus and the conditions for virus propagation and purification highlighted an unexpected complexity in the arrangement of the proteins forming the capsid shell.
The assembly pathway of large viruses with a double β-barrel MCP is understood, but these archaeal viruses encode for two MCPs, with a vertical single β-barrel fold, and display pseudo-hexagonal building blocks (capsomers) with different morphologies, raising the question of how this more complex self-assembly is achieved.
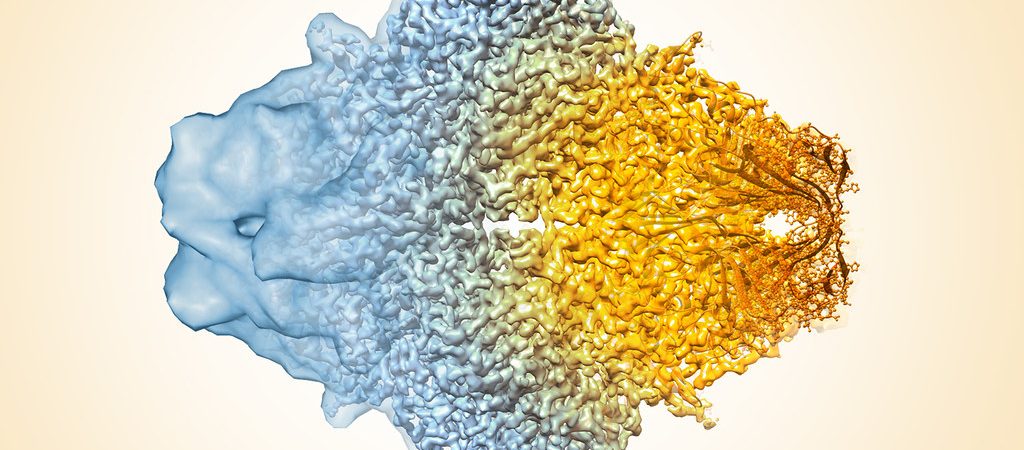
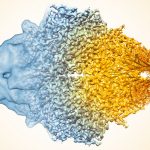
To investigate, the team used cryo-electron microscopy (cryo-EM) to determine the structure of the viruses at near-atomic resolution. They collected 2,786 cryo-images of HHIV-2 in movie format (7 frames) at the Netherlands Center for Electron Nanoscopy (NeCEN), and 3,218 HCIV-1 cryo-images in movie format (27 frames) at the electron Bio-Imaging Centre (eBIC).
Virus GPS
Together with proteomics data, structures determined by cryo-EM based provide insights into the assembly mechanism of this type of virus, and into those with membrane-less double β-barrel MCPs. As Prof Abrescia explains, “Two proteins located between the capsid shell and the outer membrane leaflet operate as a kind of GPS. These two different proteins act as navigation buoys for the direction of assembly.”
The results explored in this paper add to our understanding of how gene duplication and gene fusion events can lead to a decrease in assembly complexity and allow us to test, in computational biology, how this apparent assembly simplification leads to the encoding of more complex information in the fused MCP gene.
Source: Diamond Light Source